Research
evolutionary systems and synthetic biology of microbes
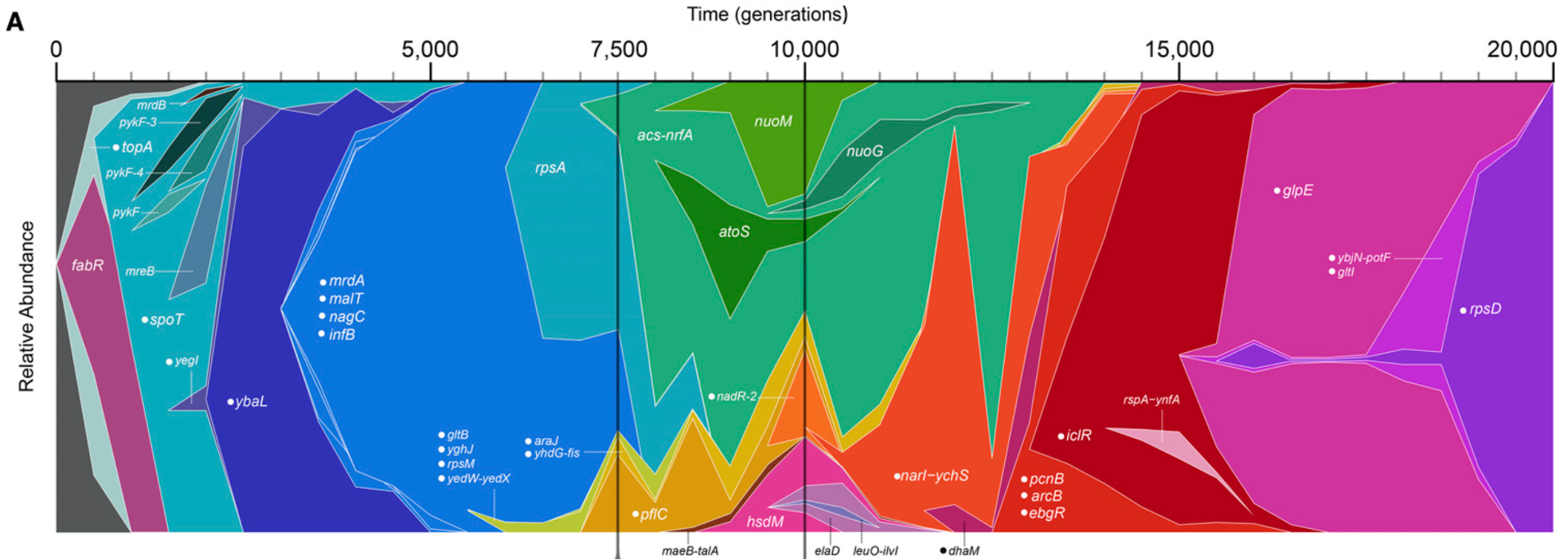
Our work focuses on molecular mechanisms that drive rapid evolution in bacteria (plasmids, mobile genetic elements, gene duplication).
To study mechanisms of microbial evolution, we use computation (data science, bioinformatics, genomics) and experiments (experimental evolution, synthetic biology) and we also do some math (equations and simulations) and machine learning. We trust data over theory, theory over hypothesis, and hypothesis over opinion. Much of our work is inspired by past and present collaborations. We have found that collaborations often open up new ideas and research directions, and often directly motivates the development of new techniques and methods.
recent work
plasmid scaling laws
We used pseudoalignment and the expectation maximization (EM) algorithm to infer plasmid copy number across all the genomes that we could get our hands on. We discovered that plasmids obey three empirical scaling laws, implying fundamental constraints on plasmid evolution and functional organization. The fundamental evolutionary and biophysical causes of these patterns remain poorly understood.
duplicated antibiotic resistance genes reveal ongoing selection and horizontal gene transfer
We discovered that duplicated antibiotic resistance genes reveal ongoing natural selection and gene transfer networks in bacteria, using mathematical modeling, synthetic biology, evolution experiments, and “big data” genomics.
publications
A current list of our publications can be found on Google Scholar.